Class Kinematic reconstruction algorithm using an Extended Kalman Filter. More...
#include <KalmanRecons.h>
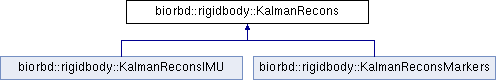
Public Member Functions | |
| KalmanRecons () | |
| Kalman reconstruction. | |
| KalmanRecons (biorbd::Model &model, unsigned int nbMeasure, KalmanParam params=KalmanParam()) | |
| Kalman reconstruction. More... | |
| virtual | ~KalmanRecons () |
| Destroy class properly. | |
| void | DeepCopy (const biorbd::rigidbody::KalmanRecons &other) |
| Deep copy of Kalman reconstruction. More... | |
| void | getState (biorbd::rigidbody::GeneralizedCoordinates *Q=nullptr, biorbd::rigidbody::GeneralizedVelocity *Qdot=nullptr, biorbd::rigidbody::GeneralizedAcceleration *Qddot=nullptr) |
| Get the state (Q, Qdot, Qddot) More... | |
| void | setInitState (const biorbd::rigidbody::GeneralizedCoordinates *Q=nullptr, const biorbd::rigidbody::GeneralizedVelocity *Qdot=nullptr, const biorbd::rigidbody::GeneralizedAcceleration *Qddot=nullptr) |
| Set the initial guess of the reconstruction. More... | |
| virtual void | reconstructFrame ()=0 |
| Proceed to one iteration of the Kalman filter. | |
Protected Member Functions | |
| virtual void | initialize () |
| Initialization of the filter. | |
| biorbd::utils::Matrix | evolutionMatrix (const unsigned int m, unsigned int n, double Te) |
| Create the evolution matrix. More... | |
| biorbd::utils::Matrix | processNoiseMatrix (const unsigned int nbQ, double Te) |
| Process the noise matrix. More... | |
| biorbd::utils::Matrix | measurementNoiseMatrix (const unsigned int nbT, double val) |
| Matrix of the noise on the measurements. More... | |
| biorbd::utils::Matrix | initCovariance (const unsigned int nbQ, double val) |
| Returns a initialized covianriance matrix. More... | |
| biorbd::rigidbody::GeneralizedCoordinates | initState (const unsigned int nbQ) |
| Initialize the states. More... | |
| void | iteration (biorbd::utils::Vector measure, const biorbd::utils::Vector &projectedMeasure, const biorbd::utils::Matrix &Hessian, const std::vector< unsigned int > &occlusion=std::vector< unsigned int >()) |
| Compute an iteration of the Kalman filter. More... | |
| virtual void | manageOcclusionDuringIteration (biorbd::utils::Matrix &InvTp, biorbd::utils::Vector &measure, const std::vector< unsigned int > &occlusion) |
| Manage the occlusion during the iteration. More... | |
Protected Attributes | |
| std::shared_ptr< KalmanParam > | m_params |
| The parameters of the Kalman filter. | |
| std::shared_ptr< double > | m_Te |
| Inherent parameter to the frequency. | |
| std::shared_ptr< unsigned int > | m_nbDof |
| Number of states. | |
| std::shared_ptr< unsigned int > | m_nMeasure |
| Number of measurements. | |
| std::shared_ptr< biorbd::utils::Vector > | m_xp |
| State vector. | |
| std::shared_ptr< biorbd::utils::Matrix > | m_A |
| Evolution matrix. | |
| std::shared_ptr< biorbd::utils::Matrix > | m_Q |
| Noise matrix. | |
| std::shared_ptr< biorbd::utils::Matrix > | m_R |
| Matrix of the noise on the measurements. | |
| std::shared_ptr< biorbd::utils::Matrix > | m_Pp |
| Covariance matrix. | |
Detailed Description
Class Kinematic reconstruction algorithm using an Extended Kalman Filter.
Definition at line 62 of file KalmanRecons.h.
Constructor & Destructor Documentation
◆ KalmanRecons()
| biorbd::rigidbody::KalmanRecons::KalmanRecons | ( | biorbd::Model & | model, |
| unsigned int | nbMeasure, | ||
| KalmanParam | params = KalmanParam() |
||
| ) |
Kalman reconstruction.
- Parameters
-
model The joint model nbMeasure The number of measurements params The Kalman filter parameters
Definition at line 25 of file KalmanRecons.cpp.
Member Function Documentation
◆ DeepCopy()
| void biorbd::rigidbody::KalmanRecons::DeepCopy | ( | const biorbd::rigidbody::KalmanRecons & | other | ) |
Deep copy of Kalman reconstruction.
- Parameters
-
other The Kalman reconstruction to copy
Definition at line 46 of file KalmanRecons.cpp.
◆ evolutionMatrix()
|
protected |
Create the evolution matrix.
- Parameters
-
m The number of degrees of freedom n The order of the Taylor development Te Is equal to 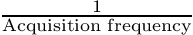
- Returns
- The evolution matrix assuming constant frame rate
Definition at line 106 of file KalmanRecons.cpp.
◆ getState()
| void biorbd::rigidbody::KalmanRecons::getState | ( | biorbd::rigidbody::GeneralizedCoordinates * | Q = nullptr, |
| biorbd::rigidbody::GeneralizedVelocity * | Qdot = nullptr, |
||
| biorbd::rigidbody::GeneralizedAcceleration * | Qddot = nullptr |
||
| ) |
Get the state (Q, Qdot, Qddot)
- Parameters
-
Q The generalized coordinates Qdot The generalized velocities Qddot The generalized accelerations
Definition at line 90 of file KalmanRecons.cpp.
◆ initCovariance()
|
protected |
Returns a initialized covianriance matrix.
- Parameters
-
nbQ The number of degrees-of-freedom val The initial value to fill the matrix with
- Returns
- The initial covariance matrix
Definition at line 208 of file KalmanRecons.cpp.
◆ initState()
|
protected |
Initialize the states.
- Parameters
-
nbQ The number of degrees-of-freedom
- Returns
- The initialized states
Definition at line 188 of file KalmanRecons.cpp.
◆ iteration()
|
protected |
Compute an iteration of the Kalman filter.
- Parameters
-
measure The vector actual measurement to track projectedMeasure The projected measurement from the update step of the filter Hessian The hessian matrix occlusion The vector where occlusionsoccurs
Definition at line 59 of file KalmanRecons.cpp.
◆ manageOcclusionDuringIteration()
|
protectedvirtual |
Manage the occlusion during the iteration.
- Parameters
-
InvTp The inverse of the Tp matrix measure The vector actual measurement to track occlusion The vector where occlusions occurs
Reimplemented in biorbd::rigidbody::KalmanReconsMarkers, and biorbd::rigidbody::KalmanReconsIMU.
Definition at line 79 of file KalmanRecons.cpp.
◆ measurementNoiseMatrix()
|
protected |
Matrix of the noise on the measurements.
- Parameters
-
nbT The number of measurements val The noise level
- Returns
- The matrix of the noise on the measurements
Definition at line 179 of file KalmanRecons.cpp.
◆ processNoiseMatrix()
|
protected |
Process the noise matrix.
- Parameters
-
nbQ The number of degrees-of-freedom Te Is equal to 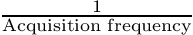
- Returns
- The noise matrix
Definition at line 130 of file KalmanRecons.cpp.
◆ setInitState()
| void biorbd::rigidbody::KalmanRecons::setInitState | ( | const biorbd::rigidbody::GeneralizedCoordinates * | Q = nullptr, |
| const biorbd::rigidbody::GeneralizedVelocity * | Qdot = nullptr, |
||
| const biorbd::rigidbody::GeneralizedAcceleration * | Qddot = nullptr |
||
| ) |
Set the initial guess of the reconstruction.
- Parameters
-
Q The generalized coordinates Qdot The generalized velocities Qddot The generalized accelerations
Definition at line 193 of file KalmanRecons.cpp.
The documentation for this class was generated from the following files:
 1.8.18
1.8.18